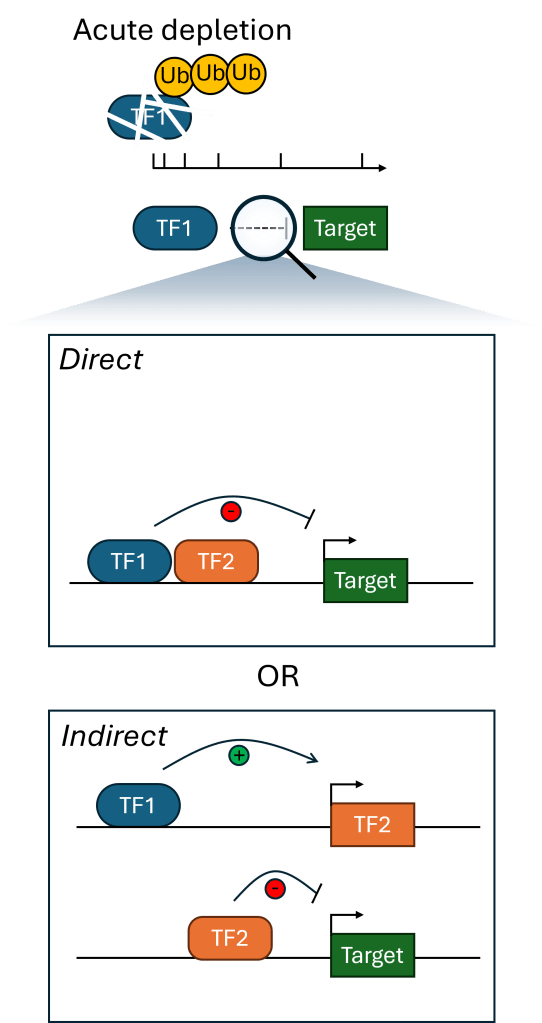
Resolving precise TF functions
Transcription factors are densely connected in gene regulatory networks, with feedback loops and regulatory cascades. This makes it challenging to identify exactly what molecular and cellular functions a TF is performing. Recent advances in chemically-inducible degron and genome editing technologies have enabled acute perturbations in sub-hour time scales. We are using these approaches with epigenomic assays to resolve direct from indirect TF functions and which key molecular events drive cellular phenotypes.
Dissecting TF disordered regions
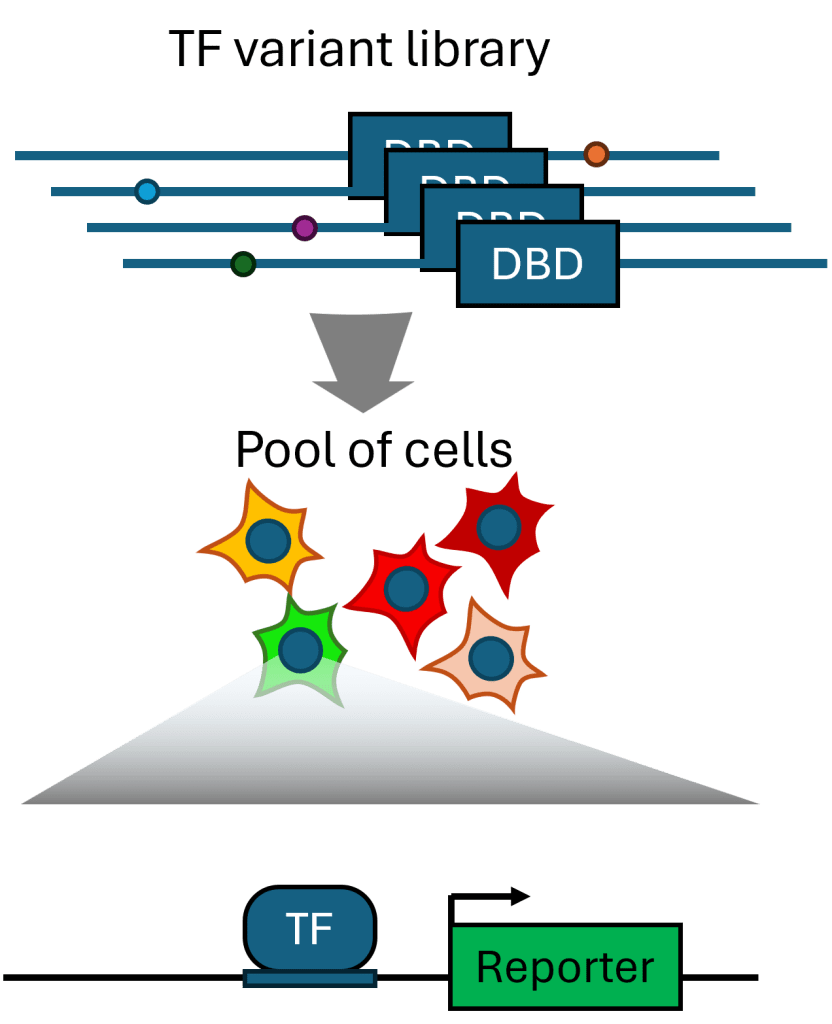
Transcription factors are typically primarily defined by their well-structured and conserved DNA-binding domains and the sequence motifs they recognize. However, paralogous TFs with highly similar DNA-binding domains (such as the large family of homeodomain TFs) can perform distinct functions, but remaining coding sequences within TFs are often poorly understood and difficult to work with biochemically. We are using high-throughput cellular genetics approaches to dissect these protein regions and learn new principles governing their activity.
Neural crest patterning and cell fates
The neural crest is considered to be the “fourth germ layer” of embryonic development in vertebrates, with important roles in the evolution of morphological diversity and in numerous diseases called “neurocristopathies”. We are using stem cell models of neural crest cell development and differentiation to better understand how they become patterned and select cell fates, and how this lineage history influences the properties of their cellular derivatives.